When working with engineering customers, a typical ETL scenario is reading in some periodically incoming data files that are in a CSV or Excel format, and running them through an ETL pipeline. The data might be manually edited by a customer at some point, or it might come automatically from some other source. While having the data streamed as something else than static files would probably be better, you often don’t get to choose the method of delivery. So you need a way to efficiently load in a bunch of files, extract the data in them, do some transformations and finally store the end result to a database.
DataFrame usecases
Now, when working with Python as we do, the most convenient option for this kind of data processing is to use a dataframe library like Pandas or Polars. A dataframe is a datastructure with columns and rows, so representing CSV data with it is quite natural. For the practical purposes of this blogpost, I will give examples using the Polars library. I personally think it’s more efficient and intuitive to use than Pandas, but the same principles I will mention apply to most dataframe related work. Using dataframes in this scenario separates the transformation from extraction. You use a similar interface to do the data transformation, not depending on the format of the original data, because the dataframe format will stay the same. Thus, if you want to switch from Excel to CSV, you might get away with as little as changing the function that is used to load the data.
In practical terms, to read in a Polars dataframe from Excel:
pl.read_excel(
source="data.xlsx",
sheet_name="data",
)
And to switch to reading a CSV:
pl.read_csv(
"data.csv",
has_header=True
)
With the same data, both will result into you having an identical dataframe to work with:
┌────────────┬──────────────┐
│ snake_name ┆ snake_length │
│ --- ┆ --- │
│ str ┆ i64 │
╞════════════╪══════════════╡
│ python ┆ 420 │
│ anaconda ┆ 370 │
│ python ┆ 350 │
│ anaconda ┆ 520 │
└────────────┴──────────────┘
Common dataframe processing pitfalls
Alright, separation of concern is nice and all, but how to best make use of the dataframe you have to do the actual T part of the ETL?
Not utilizing vectorized operations
Well, the first thing I notice in a poorly written dataframe codebase is the use of loops instead of vectorized operations. Rather than iterating through individual elements with Python loops, you should be taking use of the underlying low-level implementation of your dataframe library to perform computations on the whole dataframe at once. So, to take full use of the rust behind Polars, take a look at these examples:
Looping through the dataframe rows:
def total_python_length(df):
result = 0
for row in df.iter_rows(named=True):
if row["snake_name"] == "python":
result += row["snake_length"]
return result
The same with polars vectorized operations:
def total_python_length(df):
return (
df.filter(
pl.col("snake_name") == "python"
)
.select("snake_length")
.sum()
.item()
)
The first snippet uses traditional Python loops and conditionals to find out the total length of all the snakes with the name “python”. The second snippet does exactly the same, but using Polars filter() and select() to do the job. The first example was roughly 10x slower than the second. When processing large volumes of data, it tends to make a difference.
Using UDF:s when a select would suffice
A similar but a bit less severe variation to looping through the dataframe is using map_rows (or apply in Pandas) in cases where it is not needed at all. Map_rows takes a custom function (UDF, user defined function) and applies to each row in the dataframe. Most of the time, you don’t actually need it.
result = df.map_rows(lambda row: row[1] * 10)
Regular select operation in this case is ~40% faster.
result = df.select(pl.col("snake_length") * 10)
Not doing schema validation
Assuming that the received data is clean as-is and does not require any schema validation before saving to a database is a big pitfall. Your code might execute fine for a long time, but one day you receive a mysterious error because someone changed the original data very subtly. Take this example: here you can see two very similar looking Excel files. See the difference?
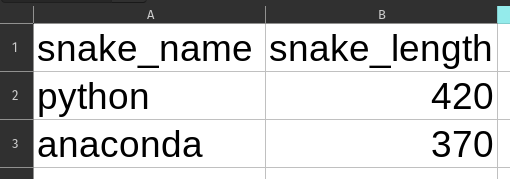
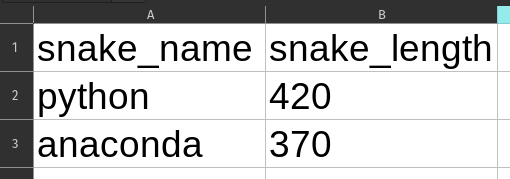
Yeah, the first one has snake length as a number and the second one as a string. Quite common problem when people copy-paste data around from different places. What happens when you try to load these to a dataframe?
For the first one
>>> print(df.dtypes)
[String, Int64]
And the second:
>>> print(df.dtypes)
[String, String]
Oopsie. That won’t end well with arithmetic operations.
One remedy is to do explicit casting…
df = df.with_columns(
pl.col("snake_length").cast(pl.Int64, strict=False),
)
… but it’s usually better to be explicit with your schema from the start.
df = pl.read_excel("data.xlsx",
sheet_name="data",
schema_overrides={"snake_length": pl.Int64}
)
You can’t alway do this, but it’s a good start. With schema_overrides, polars will try to convert your data to the defined type, even when the original is something else. If there are invalid datapoints (say, 370cm instead of 370 in the column), it will silently convert those cells to null.
Assuming the sorting order
As with schemas, try to be explicit with sorting order too, especially when presenting the data to a human. Let’s work with this dataframe, all fine and dandy and sorted by timestamp:
┌─────────────────────┬────────────┬──────────────┐
│ timestamp ┆ snake_name ┆ snake_length │
│ --- ┆ --- ┆ --- │
│ datetime[ms] ┆ str ┆ i64 │
╞═════════════════════╪════════════╪══════════════╡
│ 2025-01-01 13:00:00 ┆ python ┆ 420 │
│ 2025-01-01 13:05:00 ┆ python ┆ 360 │
│ 2025-01-01 13:15:00 ┆ anaconda ┆ 390 │
│ 2025-01-01 14:20:00 ┆ anaconda ┆ 370 │
│ 2025-01-01 14:45:00 ┆ python ┆ 410 │
└─────────────────────┴────────────┴──────────────┘
Now let’s do some grouping by timestamp hour and snake_name:
df.group_by(
pl.col("timestamp").dt.hour().alias("hour"),
"snake_name"
).agg(
pl.col("snake_length").mean().alias("mean_length"),
)
┌──────┬────────────┬─────────────┐
│ hour ┆ snake_name ┆ mean_length │
│ --- ┆ --- ┆ --- │
│ i8 ┆ str ┆ f64 │
╞══════╪════════════╪═════════════╡
│ 13 ┆ python ┆ 390.0 │
│ 14 ┆ python ┆ 410.0 │
│ 14 ┆ anaconda ┆ 370.0 │
│ 13 ┆ anaconda ┆ 390.0 │
└──────┴────────────┴─────────────┘
Notice how we are not sorted by the first column any more. Polars group_by does not maintain order by default, so let’s try that first:
df.group_by(
pl.col("timestamp").dt.hour().alias("hour"),
"snake_name",
maintain_order=True
).agg(
pl.col("snake_length").mean().alias("mean_length"),
)
┌──────┬────────────┬─────────────┐
│ hour ┆ snake_name ┆ mean_length │
│ --- ┆ --- ┆ --- │
│ i8 ┆ str ┆ f64 │
╞══════╪════════════╪═════════════╡
│ 13 ┆ python ┆ 390.0 │
│ 13 ┆ anaconda ┆ 390.0 │
│ 14 ┆ anaconda ┆ 370.0 │
│ 14 ┆ python ┆ 410.0 │
└──────┴────────────┴─────────────┘
Somewhat better, but not very consistent still. There’s also a gotcha: this makes group_by slower and blocks the using of streaming engine, which usually allows you to operate on data larger than your RAM by using for example scan_csv instead of read_csv.
It’s always best to be exlplicit with your sorting order, and (unless otherwise required) do that in the end when presenting the data to a human:
df.group_by(
pl.col("timestamp").dt.hour().alias("hour"),
"snake_name"
).agg(
pl.col("snake_length").mean().alias("mean_length"),
).sort("hour", "mean_length")
┌──────┬────────────┬─────────────┐
│ hour ┆ snake_name ┆ mean_length │
│ --- ┆ --- ┆ --- │
│ i8 ┆ str ┆ f64 │
╞══════╪════════════╪═════════════╡
│ 13 ┆ anaconda ┆ 390.0 │
│ 13 ┆ python ┆ 390.0 │
│ 14 ┆ anaconda ┆ 370.0 │
│ 14 ┆ python ┆ 410.0 │
└──────┴────────────┴─────────────┘
Assuming sorting order also becomes a problem with another antipattern: doing manual concatenations instead of using join, but that’s a whole another story.
Want to know more?
Dataframes have a lot of gotcha’s, but they are a powerful tool when used correctly. If you want help with processing large datasets stored as CSV or Excel etc., or want learn how to optimize your existing dataframe implementation, feel free to contact us. Interjektio has worked with these kinds of usecases with several customers before, so we know how to work around the pitfalls.